Intensified DoE and hybrid modeling accelerate upstream process characterization
Process characterization is necessary for the biopharmaceutical industry to guarantee a robust and uniform product quality output. Typically, two elements are applied to accomplish this task:
- statistical design of experiments (DoE)
- process modeling
We already gave detailed insights into the need and procedure for this step during upstream processing and highlighted hybrid modeling to be the most suitable model structure. In summary, cultivations in this generated design space must be performed to characterize and describe it. Herein, often a large number of required experiments is a laborious, time-consuming issue which remains to be solved.
“How can process characterization be accelerated?”
Now, to reduce this experimental workload, we present intensified design of experiments (iDoE), i.e., intra‐experimental shifts of critical process parameters (CPP). Hereby, more than one of these CPP combination settings of the design space can be characterized per experiment, leading to a more rapidly screened design space. In our particular case, we performed two of these shifts per experiment, resulting in three instead of one characterized CPP combination setting per experiment.

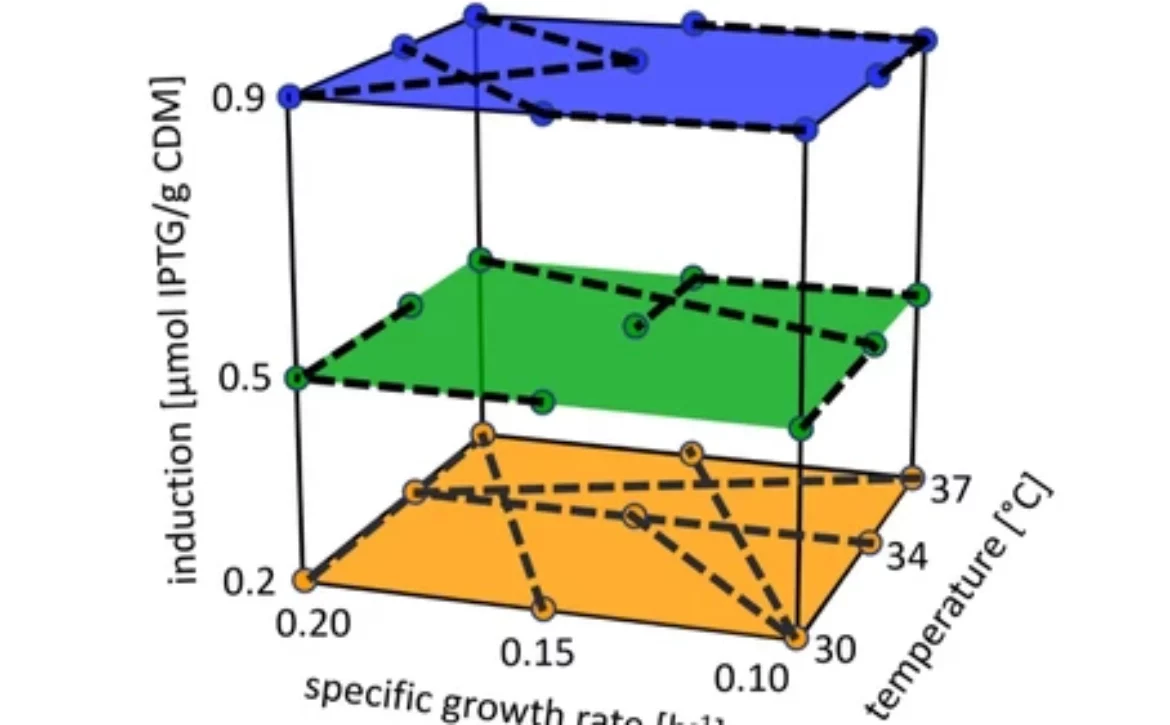
schematic operation procedure intensified design of experiments intra-experimental setpoint shifts
Process operation of our intensified E. coli fed-batch cultivations. After the batch-Glucose is consumed, the feed starts. The induction of the cells for protein production takes place after the first out of four calculated doublings (generations). After each following generation, an intra-experimental shift to a different CPP combination setting is performed, resulting in three instead of typically only one characterized setting per cultivation.
We demonstrate that by utilizing iDoE for a full factorial design space characterization, the experimental workload is reduced by more than 66%, i.e., 9 instead of 27 experiments, saving time, raw materials and goods.

Comparison of a classical static DoE and the respective iDoE approach with three CPPs, each at three levels. The full factorial design space characterization requires 27 experiments using DoE (left panel), one for each CPP combination setting while the iDoE (right panel), containing intra-experimental CPP shifts (dotted black lines), enables the same full factorial characterization with only 9 experiments.
“Are iDoE results applicable for process modeling?”
To prove the functionality of our iDoE setup, we
- characterized the complete design space from a previous study using iDoE
- trained a hybrid model on these intensified data
- and evaluated and compared the performance of both hybrid models, either trained on static DoE or iDoE
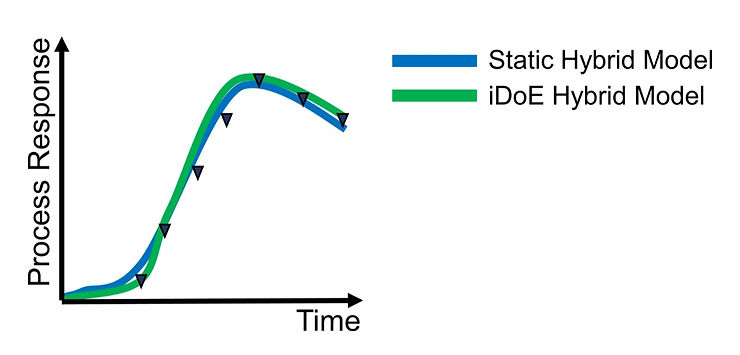
The exemplary performance of the hybrid models on predicting a process response. The process response (black triangles) over the cultivation time, e.g., the product titer, is accurately predicted by the static (solid blue line) and iDoE hybrid model (solid green line) with similar performance.
Not only were we able to reduce the number of required experiments by two thirds but moreover, the iDoE hybrid model performs with a similar accuracy compared to the static hybrid model, simultaneously predicting the biomass concentration and product titer. This highlights the potential of this combinatorial approach to accelerate bioprocess characterization.
We hope this quick excursion into the world of upstream & process modeling, utilizing novel and promising techniques awakens your interest in this topic. For more detailed information check out our publication.
Sincerely,
Novasign team