Reduced experimental burden in CHO process development across scale: hybrid modeling and iDoE
Process development in a QbD approach is accompanied by high experimental effort due to a design of experiments (DoE) approach at different scales, consuming large quantities of raw materials and time. For upstream process development of mammalian cell culture, this procedure is highly exhausting because of the more complex media, potentially expanding the design space and the slower growth rates compared to microbial systems. To reduce this experimental burden and accelerate upstream process development, we emphasize:
- hybrid process modeling
- in combination with intensified design of experiments (iDoE)
Hybrid modeling increases process understanding by providing a time-resolved relationship between critical process parameters and key process variables of interest, such as the cell concentration and product titer. iDoE provides more process response information in less overall process time by applying intra-experimental parameter shifts Thereby iDoE enables process characterization of more than just one parameter set combination in the chosen design space. A combination of hybrid modeling and iDoE renders beneficial, especially at larger scales at which costs per experiment are tremendous. Therefore, to save valuable resources, profound process knowledge should already be gathered at a small scale and only complemented with a few validation experiments on a larger scale. However, the combination of iDoE and hybrid modeling approach for mammalian cell culture was not reported so far.
‘Shake flask experiments for estimating key variables at the bioreactor scale?’
To shed light on this promising combination to save resources and time, Bilfinger and Novasign joined forces, investigating the transferability of process knowledge packed into hybrid models along process scales as well as identifying hurdles and challenges in a CHO bioprocess. Therefore, a two-dimensional design space (three levels for the cultivation temperature and the glucose concentration in the feed) was completely characterized in bolus fed shake flask duplicates. For these cultivations, necessary factors in developing a hybrid model estimating the viable cell concentration and product titer were identified, e.g., critical process parameters and amino acid consumption pattern. For the modeling part, we used our Hybrid Modeling Toolbox. The performance of the shake flask hybrid model was assessed on 15 L continuous fed stirred tank bioreactor cultivations (1:50) within the same design space, performed with static (common) and intensified process settings. An exemplary presentation of the hybrid model performance on estimating the two key process variables in a 15 L iDoE cultivation is presented in Figure 1.
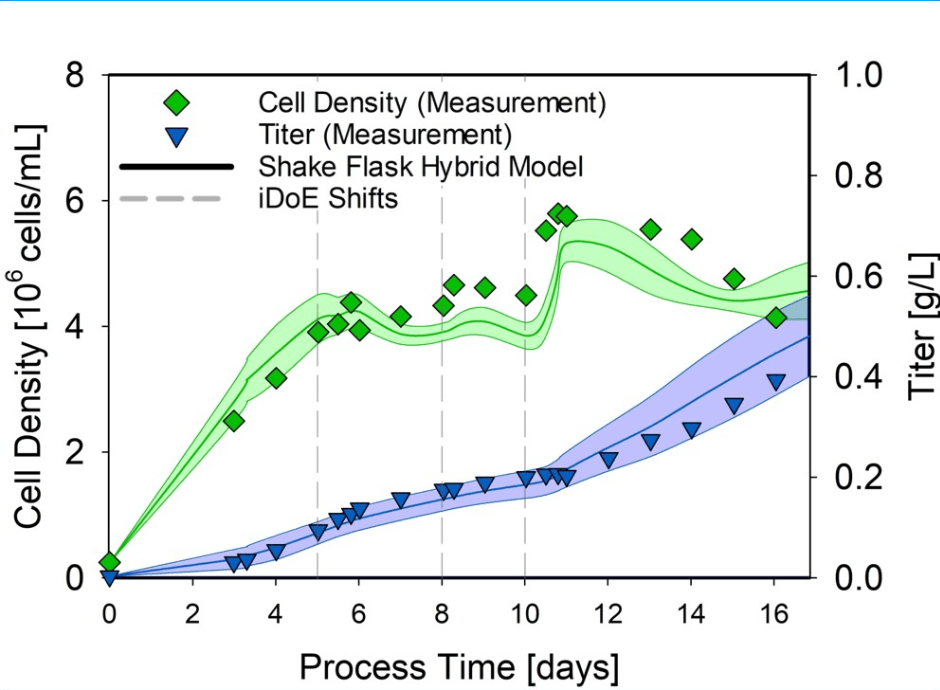
Figure 1. Shake flask hybrid model transferred to 15 L iDoE cultivation to assess model predictions. Green and blue lines indicate changes in the cell density and normalized titers, respectively.
Following the presented cultivation, the overall shaker-scale hybrid model performance proved suitable for transferring the established model structure to the 15 L scale, estimating the viable cell concentration and the product titer with an NRMSE of 10.92 % and 17.79 %, respectively.
‘How applicable is the iDoE concept for mammalian bioprocesses?‘
After proving the transferability of the shake flask hybrid model to the 15 L scale, we were also interested in the applicability of using mammalian iDoE bioprocesses for training such a hybrid model, which would enable accelerated process characterization in future studies when planning iDoE accordingly. While for microbial processes the general applicability has been proven, the applicability of iDoE for mammalian cell cultures remains to be answered. Especially the required time to see a cellular response after a process parameter shift was of high interest. To investigate iDoE model performance, another hybrid model was developed based on the 15 L iDoE cultivations with the same structure as the previous shake flask hybrid model, and its performance was tested on the static 15 L cultivations (Figure 2).
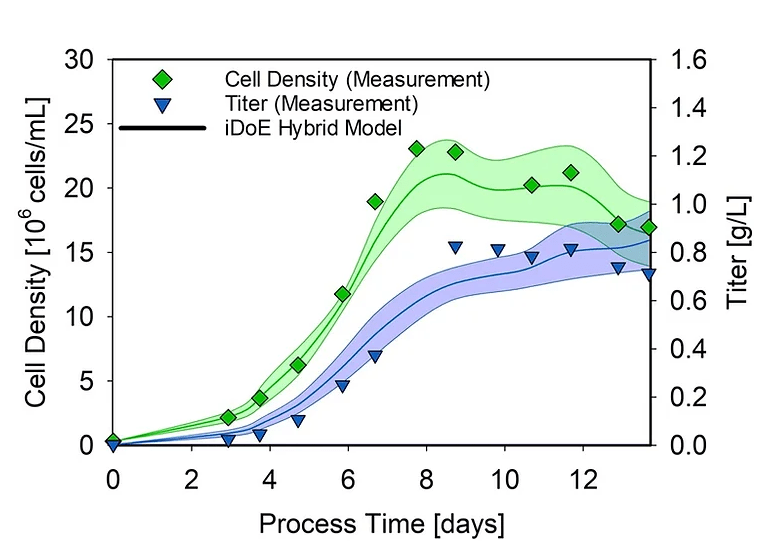
Figure 2. Hybrid model trained on iDoE data estimating key process variables in a static 15 L DoE cultivation. Green and blue lines indicate changes in the cell density and normalized titers, respectively.
The iDoE hybrid model overall performed comparable to the previous hybrid model, displaying NRMSE values of 13.75 % (viable cell concentration) and 21.13 % (product titer). These results demonstrate the suitability and potential of the combined approach for mammalian bioprocesses highlighted by good predictions errors when transferring the models trained on iDoE to DoE cultivations.
During this project collaboration, we were able to gain many insights and findings, which will be used to build further knowledge. We could demonstrate
- the transferability of process knowledge by hybrid modeling from 300 mL bolus feeding shake flask experiments to a 15 L continuous feeding stirred tank bioreactor within the same design space, requiring only minimal recalibration.
- the general applicability of iDoE for mammalian bioprocesses enabling faster process characterization to cover a particular design space with fewer experiments. For future studies, we will focus on maximum suitable process changes and the ideal time point of each iDoE cultivation shift to maximize the learning process even further.
The detailed story and conclusion can be found in our open access publication.
Contact us if you are also interested in using hybrid modeling and iDoE during your process development and don’t know how and where to start.